Allergen-specific immunotherapy (AIT) is the one therapy of IgE-mediated allergy symptoms to this point that has a sustained impact on scientific signs and might modify the course of the illness. It’s an allergen-specific therapy and due to this fact requires the right identification of the disease-causing allergens.
Moreover, AIT is a time-consuming therapy for which the efficacy depends on a number of components. Due to this fact, diagnostic exams and biomarkers are wanted that facilitate (1) number of the right allergens based on the affected person’s particular person sensitization profile and (2) to watch the consequences of AIT.
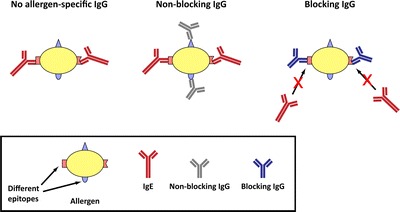
This could present help for the choice to proceed, modify, or discontinue vaccination. One vital mechanism of motion of AIT is the induction of allergen-specific antibodies that compete with IgE for the binding to allergen molecules, therefore known as blocking antibodies.
It was proven in a number of research that the induction of blocking antibodies by AIT, and their specificity will be measured by allergen microarrays. Inhibition of allergen-specific IgE binding by blocking antibodies can be decided by microarrays and is related to adjustments in scientific parameters or different in vivo and in vitro assays demonstrating efficacy of AIT.
Moreover, allergen microarrays enable dedication of IgE sensitizations in direction of a complete set of allergen molecules and due to this fact are effectively suited to figuring out the disease-causing allergens for proper prescription of AIT.
Thus, diagnostic exams based mostly on microarrayed allergens will be helpful in figuring out the right prescription of AIT and can be utilized to watch efficacy of AIT.
The impact of thermal processing on the behaviour of peanut allergen peptide targets utilized in a number of response monitoring mass spectrometry experiments.
Mass spectrometry-based strategies provide an alternate technique of figuring out allergens in meals. While focused strategies are prone to provide probably the most sturdy method for detection and quantification, little is understood about how meals processing might have an effect on the behaviour of peptide targets.
A scientific examine has been undertaken to research the consequences of thermal processing (boiling, roasting, frying) on the behaviour of a set of peanut peptide targets representing the key clinically-relevant allergens.
Initially the impact of thermal processing on protein extractability was investigated and a mass spectrometry-compatible buffer recognized comprising 50 mM Tris-HCl, pH 8.Eight containing 50 mM dithiothreitol and 0.04% (w/v) acid labile detergent which was capable of extract 45-100% of protein from uncooked, boiled, roasted and fried peanuts utilizing sonication at 60 °C.
Eight peptide targets have been recognized together with two peptides from every cupin allergen, Ara h1 and Ara h3 and 4 peptides from the prolamin superfamily allergens Ara h2, 6 and seven.
AQUA peptide requirements have been synthesised and used to undertake multiple-reaction monitoring experiments, giving assay sensitivities of 0.1-30 amoles of peptide on-column (3 : 1 sign : noise), calculated limits of quantification between 96-1343 amoles of peptide on-column and a linear dynamic vary of 4-5 orders of magnitude.
Absolute quantification of particular person peanut allergens in thermally processed samples confirmed that peptide targets within the cupin allergens have been extra susceptible to processing-induced results than these from Ara h2, 6 and seven. Targets flanked by arginine residues confirmed higher thermostability.
Identification of processing-stable targets, coupled with extra environment friendly extraction procedures and a large dynamic vary, exhibits that focused mass spectrometry strategies have nice potential as a further methodology for quantifying peanut allergens in complicated meals matrices.
